Expanding the known proteome with ribosome profiling (Ribo-seq) data
Introduction
Ribosome profiling (Ribo-seq)’s capture of the precise location and density of ribosomes across the entire transcriptome provides a snapshot of actively translated regions. Recent research has leveraged Ribo-seq’s inherent triplet periodicity signal and mRNA positional information to predict the translation of open reading frames (ORFs) alternative to the annotated protein coding ORF (CDS)1,2. Alternative translated ORFs (altORFs) include previously unannotated small ORFs (sORFs), ORFs upstream (uORFs)3 or downstream (dORFs) or overlapping4 the CDS as well as ORFs nested within the CDS5. Additionally extended proteins originating from N-terminal extensions2 or stop codon readthrough6 have been discovered with Ribo-seq.
Here, we illustrate how the EIRNA Bio Connect platform can reveal novel peptides such as uPeptides, dPeptides and other microproteins7 and thereby expand the known proteome towards better understanding of disease and development of diagnostics and therapies.

Figure 1 uORF prediction for human gene IFRD2 using algorithms implemented on the Connect platform. The sub-codon profile for the annotated IFRD2 transcript ENST00000417626 shows the CDS originating RPFs are translating in Frame 3 (coloured blue). The yellow highlighted region in the 5’leader (5’UTR) reveals that the majority of RPFs in this region are coloured red indicating that they likely originate from the translation of a non-annotated AUG-initiated uORF in Frame 1. AUG codons are represented by vertical white bars and stop codons by vertical black bars in the ORF architecture (bottom panel).

The EIRNA Bio Connect Approach: Finding novel peptides made easy
On EIRNA Bio Connect we provide a suite of algorithms to detect translation in altORFs as well as N-terminal extensions and stop codon readthrough. Our browser-based platform requires no prior bioinformatic experience to run the algorithms on your own data. Capitalizing on the triplet periodicity signal and framing bias as well as the mRNA positional information in your Ribo-seq data enables previously hidden layers of translational control to be discovered. The outputs include interactive sub-codon profiles (Figure 1) where each ribosome protected fragment (RPF) is coloured according to the reading frame it aligns to. The interactive and zoom nature of the platform reveals the sequences of the novel peptides (Figure 2). Additionally, the Connect functionality provides larger spreadsheet databases which include the amino acid sequences of all predicted novel peptides.
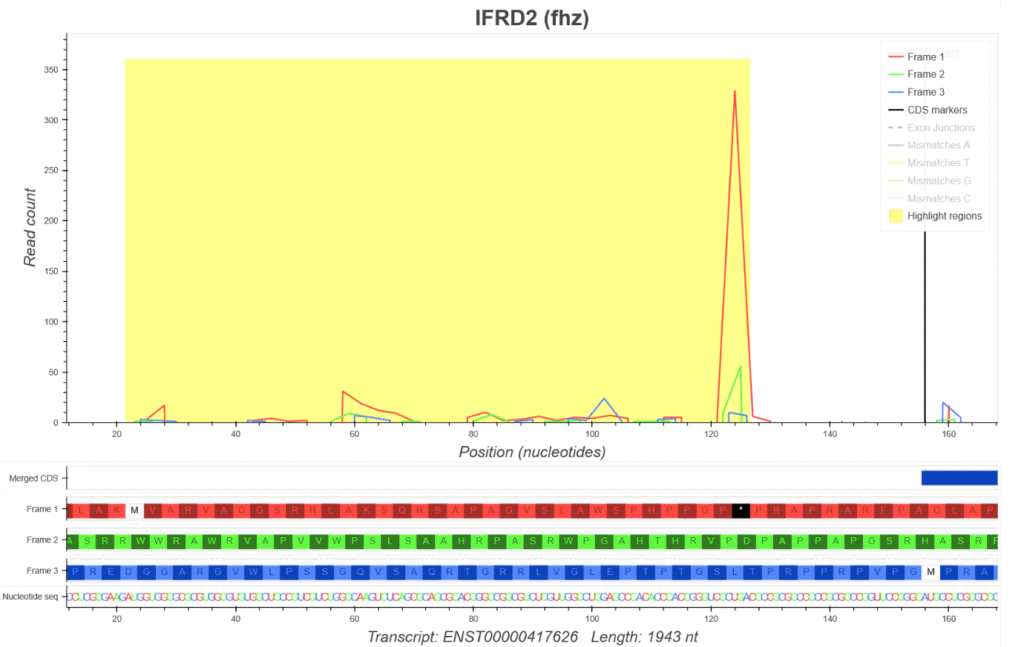
Figure 2 uPeptide prediction for human gene IFRD2 using algorithms implemented on the Connect platform. The zoomed sub-codon profile for the annotated IFRD2 transcript ENST00000417626 where the yellow highlighted region in the 5’leader (5’UTR) suggest the translation of a uORF in Frame 1 with the predicted peptide MVARVACGSRRLAKSQRSAPAGVSLAWSPHPPGP.
Applications and Implications
The discovery of alternative translation events via Ribo-seq enhances our understanding of gene regulation, proteome diversity, and potential novel functions of alternative proteins in various biological processes, including disease. Recent research illustrates how microproteins discovered using Ribo-seq can provide additional targets for immunotherapy7.
Challenges and Future Directions
A strong triplet periodicity signal is crucial for reliable altORF prediction. In addition, complex computational pipelines and algorithms are required to leverage the triplet periodicity signal for robust predictions. In EIRNA Bio, each Ribo-seq dataset goes through a series of wet-lab and bioinformatic quality control checks to maximise the triplet periodicity signal as well as sequence read coverage. The data processing is carried out by highly experienced bioinformaticians so that all you need to do is immediately start exploring for novel peptides in your own data on EIRNA Bio Connect.
Conclusion
AltORF translation refers to the process where ribosomes translate non-canonical ORFs that are different from the primary annotated coding sequences. These altORFs can produce previously unrecognized peptides and proteins, contributing to the complexity of the proteome. The discovery of alternative translation events via Ribo-seq enhances our understanding of gene regulation, proteome diversity, and potential novel functions of alternative proteins in various biological processes.
Stay tuned for how EIRNA Bio Connect can provide additional insights for your own data.
Over the course of the coming months, we will highlight additional EIRNA Bio Connect functionality to illustrate how our interactive platform can help advance your own research questions.
1. Michel AM, Roy Choudhury K, Firth AE, Ingolia NT, Atkins, JF, Baranov PV. (2012) Observation of dually decoded regions of the human genome using ribosome profiling data. Genome Res, 22:2219-2229
Fedorova AD, Kiniry SJ, Andreev DE, Mudge JM, Baranov PV. (2022) Thousands of human non-AUG extended proteoforms lack evidence of evolutionary selection among mammals. Nat Commun. 13:7910
Andreev DE, O’Connor PBF, Fahey C, Kenny EM, Terenin IM, Dmitriev SE, Cormican P, Morris DW, Shatsky IN, Baranov PV (2015) Translation of 5′ leaders is pervasive in genes resistant to eIF2 repression. Elife
Loughran G, Zhdanov AV, Mikhaylova MS, Rozov FN, Datskevich PN, Kovalchuk SI, Serebryakova MV, Kiniry SJ, Michel AM, O’Connor PBF, Papkovsky DP, Atkins JF, Baranov PV, Shatsky IN, Andreev DE. (2020) Unusually efficient CUG initiation of an overlapping reading frame in POLG mRNA yields novel protein POLGARF. Proc Natl Acad Sci U S A. 117:24936-24946
Brunet MA, Jacques JF, Nassari S, Tyzack GE, McGoldrick P , Zinman L , Jean S, Robertson J , Patani R, Roucou X. (2021) The FUS gene is dual-coding with both proteins contributing to FUS-mediated toxicity. EMBO Rep Jan 7;22(1):e50640
Yordanova MM, Loughran G, Zhdanov AM, Mariotti M, Kiniry SJ, O’Connor PBF, Andreev DE, Tzani I, Saffert P, Michel AM, Gladyshev VN, Papkovsky DB, Atkins JF, Baranov PV. (2018) AMD1 mRNA employs ribosome stalling as a mechanism for molecular memory formation. Nature 553:356-360
Camarena ME, Theunissen P, Ruiz M, Ruiz-Orera J, Calvo-Serra B, Castelo R, Castro C, Sarobe P, Fortes P, Perera-Bel J, Mar Albà M. (2024)Microproteins encoded by noncanonical ORFs are a major source of tumor-specific antigens in a liver cancer patient meta-cohort. Sci Adv Jul 12;10(28):eadn3628.



