
When it comes to early-stage drug discovery and preclinical development, understanding the complexities of translational regulation is a persistent challenge. Traditional transcriptomics methods, such as RNA-seq, provide insights into mRNA abundance but deliver no information on the RNA translational processes that determine protein output. Polysome-Seq bridges this gap by directly linking mRNA abundances with the number of associated active ribosomes to provide a highly accurate measure of translational efficiency. This approach offers a high-resolution view of gene expression at the protein synthesis level. By unveiling translational dynamics, Polysome-Seq can transform our understanding of post-transcriptional regulation, optimize therapeutic interventions and pave the way for groundbreaking advancements in personalized medicine.
Advancing Translation Research
Polysome-Seq is an innovative approach that combines polysome profiling with next-generation RNA sequencing to analyze mRNA-ribosome interactions across the entire transcriptome. This methodology allows researchers to explore the translation status of specific mRNAs, revealing insights into ribosome loading and translational efficiency. With Polysome-Seq, researchers can dissect the molecular mechanisms underlying translational regulation, paving the way for advancements in drug development and specialised therapeutic areas, such as gene therapy and personalized medicine.

What is Polysome-Seq?
Polysome-Seq integrates polysome profiling, the separation of ribosome-bound mRNAs on a linear sucrose gradient, with high-throughput sequencing of RNA-seq libraries. As standard, this technique categorizes mRNAs into lowly translated or light (1-3 ribosomes per transcript) and highly translated or heavy (>3 ribosomes per transcript) fractions, providing a detailed view of transcripts whose translational activity shifts between experimental conditions. However, the flexibility of the approach allows for up to 18 fractions to be collected, enabling granular resolution of translational states.
Key Benefits
- Quantify shifts in translational efficiency by comparing light versus heavy polysome association across conditions.
- Resolve transcript isoform–specific translation, revealing how alternative splicing affects ribosome occupancy.
- Achieve fine-scale resolution of ribosome loading across polysome gradients using multi-fraction analysis.
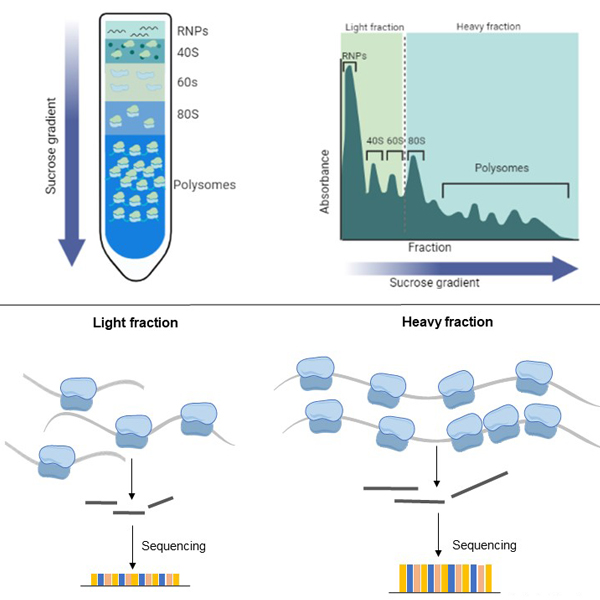
Key Features of Polysome-Seq
Quantitative Resolution
Quantifies ribosome occupancy for mRNA and noncoding RNA, distinguishing between low and high translational activity.
Optimize Codon Usage
Can be implemented on samples transfected with pooled reporter sequences in a massively paralleled manner to identify which 5'UTR, RNA secondary structure, or CDS codon sequence results in optimal ribosome loading to maximize protein expression.
Dynamic Flexibility
Offers customized fraction collection and library preparation to suit the customer’s specific research needs.
Advanced Insights
Enables exploration of post-transcriptional regulation, including translation of alternative splice variants and mRNA decay pathways.
Challenges associated with Polysome-Seq
Data Generation
Challenge 1
Technical Complexity: Polysome profiling requires precise sample preparation and fractionation expertise. Mishandling can compromise data quality, demanding skilled execution to ensure reliable, reproducible results for accurate translational analysis.
Challenge 2
High Data Volume: Disome-seq produces complex datasets with numerous data files per sample, demanding advanced computational tools and expertise to manage, integrate, and analyze the data for meaningful insights into ribosome dynamics and translational regulation.
Challenge 3
Resource Intensity: Disome profiling requires access to expensive specialized equipment and skilled personnel, making in-house setup resource-intensive and often impractical for many research laboratories.
Solution 1
Solution 1
Experience & Expertise: EIRNABio’s expert team, skilled in polysome profiling and RNA sequencing, ensures high-quality, reproducible data. Our proven expertise delivers reliable results, streamlining your research with precision and consistency across all projects.
Solution 2
State-of-the-Art Methods: We use proprietary polysome profiling techniques and precise fractionation to ensure maximum RNA yield and quality, enabling reliable data generation and robust downstream disome-seq analysis.
Solution 3
Tap into a pre-built infrastructure: We offer access to established infrastructure and expert workflows, removing the need for expensive equipment and training while delivering high-quality, reproducible disome-seq data ready for analysis.
Challenge 2
High Data Volume: Disome-seq produces complex datasets with numerous data files per sample, demanding advanced computational tools and expertise to manage, integrate, and analyze the data for meaningful insights into ribosome dynamics and translational regulation.
Solution 2
State-of-the-Art Methods: We use proprietary polysome profiling techniques and precise fractionation to ensure maximum RNA yield and quality, enabling reliable data generation and robust downstream disome-seq analysis.
Challenge 3
Resource Intensity: Disome profiling requires access to expensive specialized equipment and skilled personnel, making in-house setup resource-intensive and often impractical for many research laboratories.
Solution 3
Tap into a pre-built infrastructure: We offer access to established infrastructure and expert workflows, removing the need for expensive equipment and training while delivering high-quality, reproducible disome-seq data ready for analysis.
Data Analysis
Challenge 1
Specialized Analysis: Analyzing Polysome-Seq data is complex and demands tailored approaches. Accurate interpretation depends on aligning analysis with the specific biological question being investigated.
Challenge 2
Data Normalization: Polysome-Seq requires integrating multiple files per sample, making accurate normalization essential for comparing translational efficiency and ribosome loading across conditions.
Challenge 3
Customized mapping may be required: Tailored pipelines are often needed to address unique transcript features, enabling accurate isoform resolution or detection of exogenous transcripts for application-specific translational analysis.
Solution 1
Solution 1
Comprehensive Analysis Platform: EIRNABio Connect allows users to visualize transcript profiles, assess translational efficiency shifts across conditions, and identify coordinated changes in polysome association—thus extracting meaningful, application-specific insights.
Solution 2
Streamlined Normalization: EIRNABio Connect automates normalization across datasets, using robust statistical methods to ensure accurate ribosome loading estimates and reliable comparisons of translational efficiency between conditions.
Solution 3
Flexible Bioinformatics Support: We provide adaptable analysis pipelines and expert guidance to accommodate unique transcript features and research goals, ensuring accurate mapping, isoform resolution, and meaningful interpretation of complex datasets.
Challenge 2
Data Normalization: Polysome-Seq requires integrating multiple files per sample, making accurate normalization essential for comparing translational efficiency and ribosome loading across conditions.
Solution 2
Streamlined Normalization: EIRNABio Connect automates normalization across datasets, using robust statistical methods to ensure accurate ribosome loading estimates and reliable comparisons of translational efficiency between conditions.
Challenge 3
Customized mapping may be required: Tailored pipelines are often needed to address unique transcript features, enabling accurate isoform resolution or detection of exogenous transcripts for application-specific translational analysis.
Solution 3
Flexible Bioinformatics Support: We provide adaptable analysis pipelines and expert guidance to accommodate unique transcript features and research goals, ensuring accurate mapping, isoform resolution, and meaningful interpretation of complex datasets.

Application Areas
Global Translation Rates
Polysome-Seq is a transformative method for analysing global translation rates, providing direct insights into ribosome density and protein synthesis efficiency. This technique separates mRNAs by ribosome association and couples it with RNA sequencing to measure translation activity.
Learn More
Decoding Alternative Splicing
Polysome-Seq offers unparalleled insights into alternative splicing, uncovering how splice variants are preferentially recruited to ribosomes for translation. Learn More
Codon Optimization
Polysome-Seq provides precise insights into codon optimization, linking codon usage to translation efficiency and mRNA stability. This method identifies how codon bias influences ribosome density and protein synthesis rates, aiding the design of therapeutic constructs and biomanufacturing strategies.
Learn More
Decoding Nonsense Suppression
Polysome-Seq provides precise insights into nonsense suppression and stop codon readthrough, linking these translational events to protein synthesis and disease mechanisms.
Learn More
Polysome profiling vs Ribosome profiling
- Both polysome profiling and ribosome profiling can be used to study global translational activity, but each has distinct strengths.
- Polysome profiling measures ribosome density of each mRNA, providing a direct measurement of translational activity.
- Ribosome profiling captures positional information of ribosome footprints at the subcodon level and is more suitable for investigating alternative start codons or open reading frames.
- Ribosome profiling can reveal codon-specific regulation of gene expression during the translational elongation stage.
- The size of ribosome footprint, which is unique to ribosome profiling, may indicate differential conformation of ribosomes or the presence of disomes.
Polysome-Seq Services FAQ
What is Polysome-Seq and how does it work?
Polysome-Seq combines polysome profiling with RNA sequencing to analyze the translational state of mRNAs. It separates mRNAs based on ribosome loading and sequences them to determine translation efficiency at the transcript level.
How is Polysome-Seq different from ribosome profiling?
While ribosome profiling focuses on ribosome-protected mRNA fragments, which are limited in length (~28-30nt) and provide restricted isoform-specific insights, Polysome-Seq leverages the longer RNA-seq reads (~200-300nt) to capture isoform-specific information on translated mRNAs. This broader view allows for the detailed analysis of transcript diversity and translational states that ribosome profiling often misses.
What are the key applications of Polysome-Seq?
Polysome-Seq enables comprehensive analysis of translational efficiency and ribosome loading dynamics. It also enables the study of mRNA stability and protein synthesis control, making it invaluable for drug discovery and personalized medicine. The technique is a critical tool in translational research, bridging the gap between the underlying molecular biology and clinical applications by providing insights into how mRNAs are actively translated into proteins.
How is Polysome-Seq relevant for therapeutic development?
Polysome-Seq provides insights into gene translation by identifying which mRNAs are being actively translated, enabling study of the translation efficiency of therapeutic protein products and their regulation by translational processes. The approach can inform Advanced Therapy Medicinal Products (ATMP) development by optimizing protein expression in therapeutic contexts. In addition, Polysome-Seq can guide gene editing strategies by highlighting how specific transcript variants are translated.
What types of samples are compatible with Polysome-Seq?
Our protocols are adaptable to a wide range of samples, including cultured cells (including primary cells) and tissues, ensuring versatility in application.
How does EIRNABio support Polysome-Seq projects?
From experimental design to data analysis, our services include high-quality library preparation, sequencing, and comprehensive bioinformatics support.
How do I get started with EIRNABio’s Polysome-Seq services?
Contact us to discuss your project needs and receive a tailored solution to achieve your research objectives.



Speak to an expert!