Navigating Translation Regulation with Precision
Understanding the nuances of molecular biology is a significant challenge for early-stage drug discovery and preclinical development. Researchers grapple with complex gene expression and protein synthesis datasets to elucidate cellular mechanisms. While traditional transcriptomics (i.e., RNA-seq) and even ribosome profiling provide insights into gene expression, they fail to fully unravel the translational dynamics at play, especially those related to ribosome interactions during elongation. This limitation can lead to data being misinterpreted, creating inefficiencies in drug development and ultimately delays in getting new therapeutics to patients. The demand to characterise the translational landscape with precision is greater than ever, and with it, the need for innovative methodologies that delve deeper into the mechanisms of translation.
Disome Profiling: A New Frontier in Translation Analysis
Disome profiling, also known as disome-seq, is a cutting-edge technology that provides orthogonal support for ribosome profiling by delivering insights into ribosome collisions, a previously underexplored aspect of translation regulation. This approach captures a unique layer of translational dynamics by examining mRNA fragments protected by two ribosomes stacked together. By revealing where ribosomes pause and collide, disome-seq offers a more detailed understanding of the regulatory mechanisms controlling translational elongation. With disome profiling, researchers can identify pause sites or explore co-translational protein folding, investigating mechanisms critical to cellular homeostasis. Such insights enhance the ability to discover novel drug targets and biomarkers, ultimately accelerating the transition from bench to bedside.


What is Disome Profiling?
Disome profiling (disome-seq) targets the physical interactions between ribosomes during elongation. These disomes form when a leading ribosome pauses, causing the trailing ribosome to collide. Disome profiling provides insight into the spatial and temporal patterns of ribosome collisions, allowing researchers to understand how translational pausing is influenced by factors such as codon usage, amino acid motifs, or structural features of the nascent peptide. This level of resolution is critical for understanding how these factors influence protein folding or how ribosome collisions trigger cellular quality control mechanisms.
Key Benefits
- Detect ribosome collisions that pinpoint severe translational pausing events missed by standard ribosome profiling.
- Resolve sequence determinants of stalling at codon level resolution.
- Link translational pauses to functional outcomes, such as activation of quality control pathways.
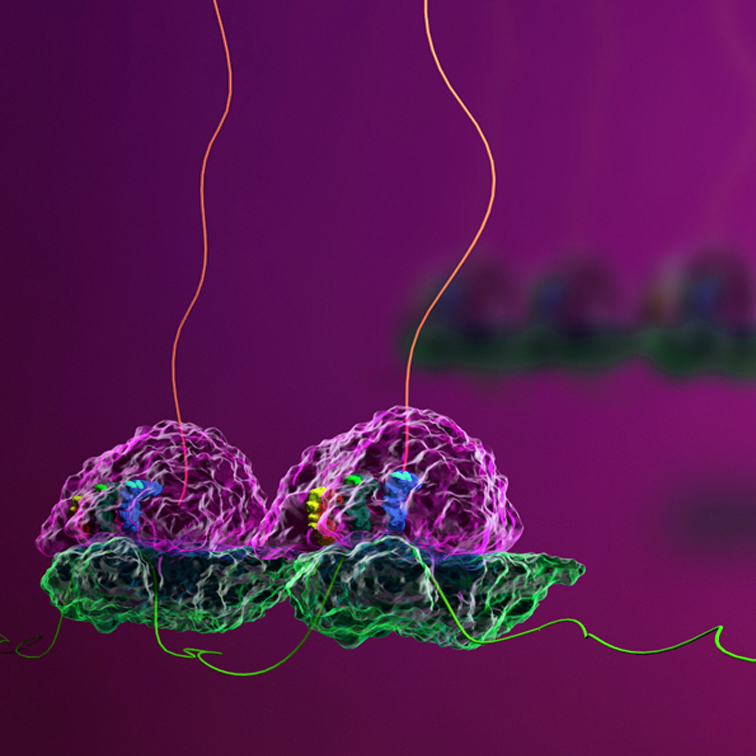
Key Features of Disome Profiling
Ribosome Collision Mapping
Disome-seq provides a transcriptome-wide view of ribosome collisions, highlighting where ribosomes pause and stack together during translation. This can reveal important regulatory sites, elucidating the source of translational bottlenecks.
Co-translational Folding Insights
Disome profiling shows how pauses contribute to the proper folding of nascent proteins, which is crucial for protein function and stability. Such insights are invaluable for therapeutic areas targeting gene translation, such as gene editing and gene therapies.
Translational Quality Control
Ribosome collisions act as triggers for activating quality control pathways, such as the ribosome-associated quality control (RQC) and stress response pathways. Identification of these collision sites aids our understanding how cells manage translational stress and maintain proteostasis.
Broad Applicability
The translational control mechanisms elucidated by Disome profiling are pertinent to a wide range of medical research fields, such as neurology and oncology. It is particularly powerful for exploring mechanisms of translational regulation, protein folding, and cellular response to environmental challenges.
Challenges Associated with Disome Profiling
Data Generation
While providing valuable insights, Disome profiling implementation is complex and presents a unique set of technical and practical challenges
Challenge 1
Technically Demanding: High-quality disome-seq demands deep expertise in molecular biology, particularly in optimizing nuclease treatment and selectively capturing disome footprints. Without mastery, troubleshooting is frequent and results can be inconsistent.
Challenge 2
Specialized Equipment & Cost: Disome profiling requires costly specialized equipment and extensive staff training, making in-house setup prohibitively expensive for many labs interested in adopting the technique.
Challenge 3
Time-consuming workflow: Disome-seq is a multi-day process requiring precision at every step—from Footprint isolation to sequencing. Any misstep can delay timelines and compromise data quality.
Solution 1
Solution 1
Domain Expertise: With decades of experience in the field of translatomics, our team delivers tailored, high-quality disome profiling services. We ensure smooth project execution and support data interpretation, helping customers extract meaningful biological insights with confidence.
Solution 2
Expertise on tap with zero set-up costs: We eliminate setup costs and risks by providing reliable, expert-led disome profiling services. With hundreds of completed projects, we are a trusted partner for generating high-quality disome-seq data.
Solution 3
Early QC to save time downstream: We apply rigorous quality control throughout the disome-seq workflow, advancing only high-quality libraries to sequencing. This ensures reliable, clean data essential for accurate interpretation and meaningful biological insight.
Challenge 2
Specialized Equipment & Cost: Disome profiling requires costly specialized equipment and extensive staff training, making in-house setup prohibitively expensive for many labs interested in adopting the technique.
Solution 2
Expertise on tap with zero set-up costs: We eliminate setup costs and risks by providing reliable, expert-led disome profiling services. With hundreds of completed projects, we are a trusted partner for generating high-quality disome-seq data.
Challenge 3
Time-consuming workflow: Disome-seq is a multi-day process requiring precision at every step—from Footprint isolation to sequencing. Any misstep can delay timelines and compromise data quality.
Solution 3
Early QC to save time downstream: We apply rigorous quality control throughout the disome-seq workflow, advancing only high-quality libraries to sequencing. This ensures reliable, clean data essential for accurate interpretation and meaningful biological insight.
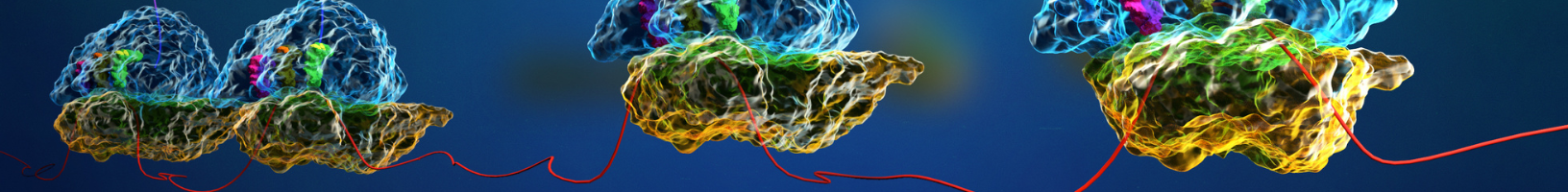
Data Analysis
Challenge 1
Solution 1
Disome profiling generates rich datasets that provide a window into translational phenomena such as ribosome pausing and ribosome quality control mechanisms. However, the analysis of disome footprints is a highly specialised area of research and can be daunting to those who are new to the field.
Challenge 1
Complex Analysis: Disome profiling reveals complex translational phenomena like ribosome pausing and quality control, but analysing disome footprints is highly specialised and can be overwhelming for researchers unfamiliar with this niche area.
Challenge 2
Not easy for beginners: Disome profiling can be overwhelming for newcomers, as interpreting complex datasets related to ribosome interactions and translational regulation requires deep domain knowledge and clear guidance on where and how to begin analysis.
Challenge 3
Understanding the context: Ribosome collisions are dynamic and influenced by biological context. Observing collision sites alone is not enough—understanding their significance requires tools that reveal surrounding translational events and patterns.
Challenge 4
Difficult to Interpret: Disome-seq data is complex and nuanced, making it easy to misinterpret without experience—especially when identifying true biological pauses versus background noise or incidental ribosome collisions.
Solution 1
Introducing EIRNABio Connect, the interactive browser based all-in-one bioinformatics platform for the analysis of translatomics data, such as that generated by disome profiling. Designed to carry out most of the “heavy lifting” for experienced bioinformaticians, whilst at the same time enabling those without in-depth bioinformatics expertise to dynamically interact with and interrogate their data.
Solution 1
Easy-to-use bioinformatics platform: EIRNABio Connect streamlines disome profiling analysis with an interactive, browser-based platform that supports both expert and novice users enabling intuitive exploration of high-resolution disome-seq data.
Solution 2
Intuitive analysis: EIRNABio Connect enables researchers to explore ribosome collision data with ease, revealing insights into stalling, peptide-mediated pauses, and stress responses—helping users connect disome profiling outputs to underlying regulatory mechanisms.
Solution 3
Dynamic Visualization of Translational Context: EIRNABio Connect's interactive visualization of ribosome density compares monosome and disome distributions, revealing the biological relevance of collision events and their role in translational control.
Solution 4
Insights from Experts: We offer expert support, tutorials, workshops, and consultations to help researchers accurately interpret disome data and uncover meaningful biological insights from ribosome collision events.
Complex Analysis: Disome profiling reveals complex translational phenomena like ribosome pausing and quality control, but analysing disome footprints is highly specialised and can be overwhelming for researchers unfamiliar with this niche area.
Easy-to-use bioinformatics platform: EIRNABio Connect streamlines disome profiling analysis with an interactive, browser-based platform that supports both expert and novice users enabling intuitive exploration of high-resolution disome-seq data.
Challenge 2
Not easy for beginners: Disome profiling can be overwhelming for newcomers, as interpreting complex datasets related to ribosome interactions and translational regulation requires deep domain knowledge and clear guidance on where and how to begin analysis.
Solution 2
Intuitive analysis: EIRNABio Connect enables researchers to explore ribosome collision data with ease, revealing insights into stalling, peptide-mediated pauses, and stress responses—helping users connect disome profiling outputs to underlying regulatory mechanisms.
Challenge 3
Understanding the context: Ribosome collisions are dynamic and influenced by biological context. Observing collision sites alone is not enough—understanding their significance requires tools that reveal surrounding translational events and patterns.
Solution 3
Dynamic Visualization of Translational Context: EIRNABio Connect's interactive visualization of ribosome density compares monosome and disome distributions, revealing the biological relevance of collision events and their role in translational control.
Challenge 4
Difficult to Interpret: Disome-seq data is complex and nuanced, making it easy to misinterpret without experience—especially when identifying true biological pauses versus background noise or incidental ribosome collisions.
Solution 4
Insights from Experts: We offer expert support, tutorials, workshops, and consultations to help researchers accurately interpret disome data and uncover meaningful biological insights from ribosome collision events.


Application Areas
Ribosome Pausing
Disome profiling is a powerful technique that captures the precise moments where ribosomes pause and collide during elongation, forming disomes. By identifying these pause sites, researchers gain unprecedented insights into translational bottlenecks, protein folding dynamics, and regulatory mechanisms that are not visible using monosome-based methods.
Learn More
Ribosome Collisions
Disome profiling is the only method that captures ribosome collisions—where stacked ribosomes accumulate due to pausing or traffic on the mRNA. These events signal translational stress and initiate quality control pathways. By mapping collision sites at nucleotide resolution, disome profiling reveals how translation is disrupted in disease, drug treatment, or stress.
Cotranslational Protein Folding
Cotranslational protein folding begins as the nascent polypeptide exits the ribosome, and regulated translation speed is essential for accurate structure formation. Disome profiling maps ribosome pausing and collisions that coordinate domain folding in real time.
uORF Regulatory Activity
Disome profiling provides unique insight into upstream open reading frames (uORFs) and their regulatory impact on translation. By capturing ribosome pausing and collisions at or near uORFs, this method reveals how these elements modulate downstream protein synthesis.
RNA-Targeting Small Molecules
Disome profiling enables precise analysis of RNA-targeting small molecules by revealing their effects on translation. Whether inducing ribosome pausing, triggering collisions, or relieving translational blocks, these compounds often act through mechanisms that only disome profiling can detect.
Translational Quality Control
Translational quality control (TQC) ensures protein synthesis accuracy by recognising stalled ribosomes and resolving translational stress. Disome profiling is the most effective way to capture ribosome collisions—key signals that initiate these surveillance pathways.
Disome Profiling Services FAQ
What is disome profiling, and how does it work?
Disome profiling is a next-generation sequencing method that provides insights into ribosome collisions during translation. It captures ribosome-protected mRNA fragments where two ribosomes are stacked together, offering a detailed view of ribosome pausing and collision dynamics.
How is disome profiling different from ribosome profiling?
While ribosome profiling provides a snapshot of elongating ribosomes (or monosomes), disome profiling specifically captures instances where ribosomes collide, providing an additional layer of insight into translation elongation, pausing, and co-translational regulation. This makes it particularly powerful for investigating factors influencing protein folding and translational quality control.
What are the key applications of disome profiling?
Disome profiling is used to study co-translational protein folding, cellular stress responses, ribosome quality control, and translational regulation. It is also valuable for identifying translational pause sites that can serve as potential drug targets or biomarkers.
Can disome profiling be used to study disease mechanisms?
Yes, disome profiling is particularly useful for studying disease mechanisms that involve disrupted translation, such as those that contribute to neurodegenerative diseases, cancer, and conditions related to protein misfolding. It provides insights into how translational pausing and ribosome collisions contribute to disease pathology.
How does EIRNABio support disome profiling projects?
We provide end-to-end support, from experiment design to data analysis. Our services include generating high-quality disome-seq libraries, performing sequencing, and providing data visualization and analysis tools to help researchers make the most of their results.
What types of samples are compatible with disome profiling?
Disome profiling can be performed on a wide range of sample types, including cultured cells, tissue samples, and even challenging low-input samples. We tailor our protocols to the specific sample type to ensure optimal results.
How do I get started with EIRNABio’s disome profiling services?
Contact us to discuss your project needs. Whether you are exploring ribosome collisions in disease models or studying stress response pathways, we offer customized service options to fit your research goals.



Speak to an expert